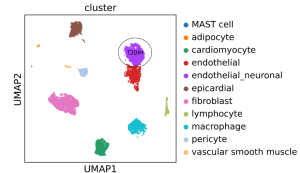
The diabetic ischemic heart (IH) is significantly associated with increased risk of heart failure and death. The pathogenesis of IH in type 2 diabetes (T2DIH) is a complex and long-lasting process, that impacts heart metabolism, leads to subclinical inflammation, oxidative stress, mitochondrial dysfunction, and cardiac cell death, potentially reshaping cardiac cell profiling. This study aims to analyse the heart transcriptome at the single-nuclei resolution of T2DIH. We performed single nucleus RNA sequencing (RNA-Seq) using 10X Genomics technology from the left atrium of: 1. patients with T2DIH; 2. non-diabetic patients with IH; and 3. normal controls (heart donors). Preliminary results in T2DIH and controls show nearly 10.000 sequenced nuclei with good quality, organized into 11 clusters of cell types, mainly composed by fibroblasts, endothelial cells, cardiomyocytes, macrophages, epicardial cells, pericytes and mast cells. Intriguingly, delineating the cellular heart landscape unveiled a distinct transcriptional mixed cluster formed by neuronal and endothelial cells only in T2DIH. These initial insights into the transcriptome profiling of the human heart at single-nuclei resolution represents a promising starting point, indicating that T2DIH have a distinct transcriptional profile. Further analyses will offer a valuable contribution to the human cell atlas in the context of T2DIH disease.