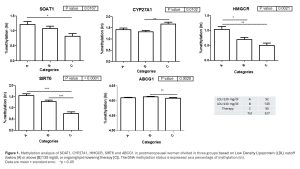
Homeostasis of cholesterol, key cardiovascular (CV) risk factor, is finely regulated. Growing evidence indicates that DNA methylation, by modifying gene expression, might participate in this process. We analyzed a possible relationship between the degree of methylation of some genes regulating cholesterol synthesis (HMGCR), accumulation (ABCG1, SOAT1 and SIRT6) and elimination (CYP27A1) and CV risk profile in 327 post-menopausal women (mean age 58.9±5.6 yrs) without previous CV events, consecutively enrolled in the CV risk screening program “Donna Cuore”. Blood chemistry, echocardiography, and carotid ultrasonography were performed, too. Participants were divided in group based on Low Density Lipoprotein (LDL) cutoff (below [A] or above [B] 130 mg/dl, or ongoing lipid-lowering therapy [C]). The methylation degree, evaluated by High Resolution Melting (a post-PCR analytical methodology used to identify methylation changes in specific regions of genes), showed a highly significant difference for some CpG sites in SOAT1 (p=0.0167), SIRT6 (p<0.0001), CYP27A1 (p=0.0102) and HMGCR (p=0.0021) genes (Fig. 1). ABCG1 was almost completely methylated in all groups. Stratifying the participants according to carotid atherosclerosis (IMT<0.9 vs increased IMT and/or presence of plaque), organ damage was associated with reduced HMGCR (p<0.0001) and increased SOAT1 (p<0.05) methylation; FSH and LH levels did not influence gene methylation. The role of epigenetics in modulating cholesterol metabolism in these women was confirmed by increased miR33a (inhibitor of SIRT6, which decreases cholesterol synthesis) and miR302a (ABCA1 modulator) in group B (p=0.008 and p=0.024, respectively). An epigenetic signature of cholesterol accumulation and homeostasis, linked to subclinical carotid atherosclerosis, is observed in aging hypercholesterolemic women.